
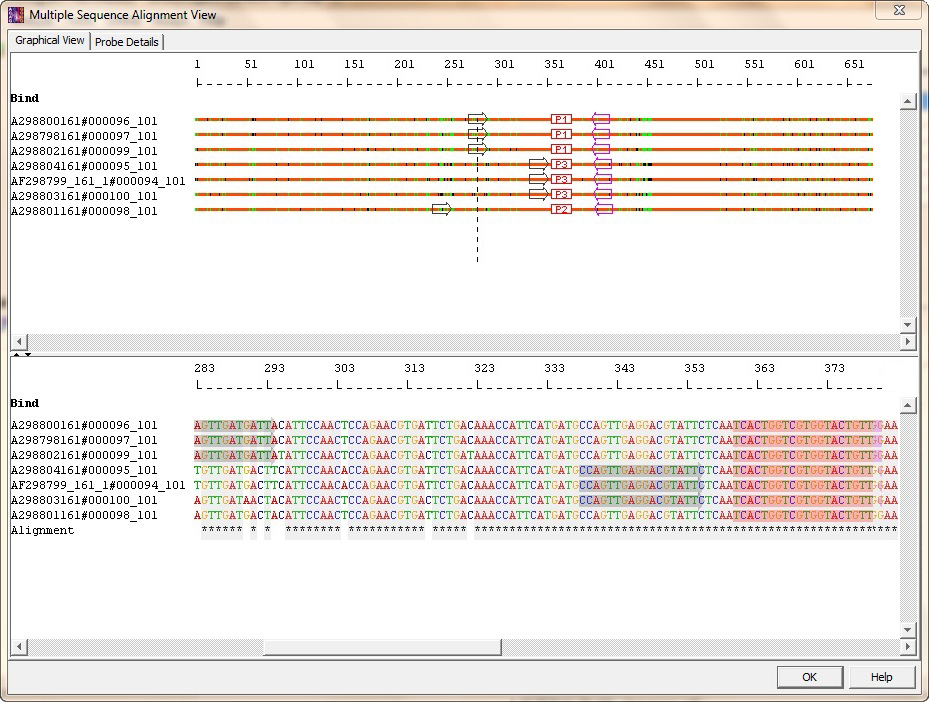
For certain species, primer pair design restricted to the conserved region was not possible. I repeated the process for the remaining four species. The primer pair was located within region conserved across the sub-species of my species of interest. AlleleID was able to design a single primer pair that would amplify only the species of interest and at the same time allow the amplification of the different strains (or sub-species) of the species in question.
They tell the program which sequences I am interested in, and which must be strictly avoided. To design a single SYBR® Green primer pair for unique identification of the first species, I made use of AlleleID's convenient 'bind to' and 'avoid' boxes. I, therefore, started by loading these alignment files, instead of using its built-in Clustal W algorithm. I had already aligned all the subspecies/strain sequences for each species before I discovered AlleleID. In more detail, my goal was to design 5 SYBR® Green primer pairs that would amplify each of the species uniquely, without amplifying the rest of the four. They have co-colonized the roots of Medicago t. The molecular work of this project depends on specific and quantitative detection of those 5 AMF species when

My PhD project aims at characterizing the functional diversity among 5 arbuscular mycorrhizal fungi (AMF) species ( Glomus intraradices, Glomus claroideum, Glomus mosseae, Gigaspora margarita, Scutellospora pellucida) in symbiosis with Medicago truncatula. AlleleID is the only tool I am aware of that can handle such oligo design challenges. I can load sequences with homologies as high as 99% and can rely on AlleleID to find species- and taxa- specific primers or probes. I have been using it frequently and am happy with it.ĪlleleID designs species and taxa identification assays, for both qPCR (TaqMan®, TaqMan® MGB, SYBR® Green and molecular beacon assays) and microarray experiments. My group purchased AlleleID, a program for designing qPCR and microarray oligos, a couple of months ago. The technique, known as real-time or quantitative PCR (qPCR), now plays a key role in scientific research. The ability to monitor the progress of PCR in real-time has revolutionized PCR-based quantification of DNA and RNA.


 0 kommentar(er)
0 kommentar(er)
